Show Advanced KnowledgeHide Advanced KnowledgeThe isoelectric point
The isoelectric point (\(pI\)) of proteins is the \(pH\) value of a solution at which the molecules positive and negative charges are in balance.
Following the definition of Brønsted-Lowry, proton donors are called acids and protein acceptors are called bases.
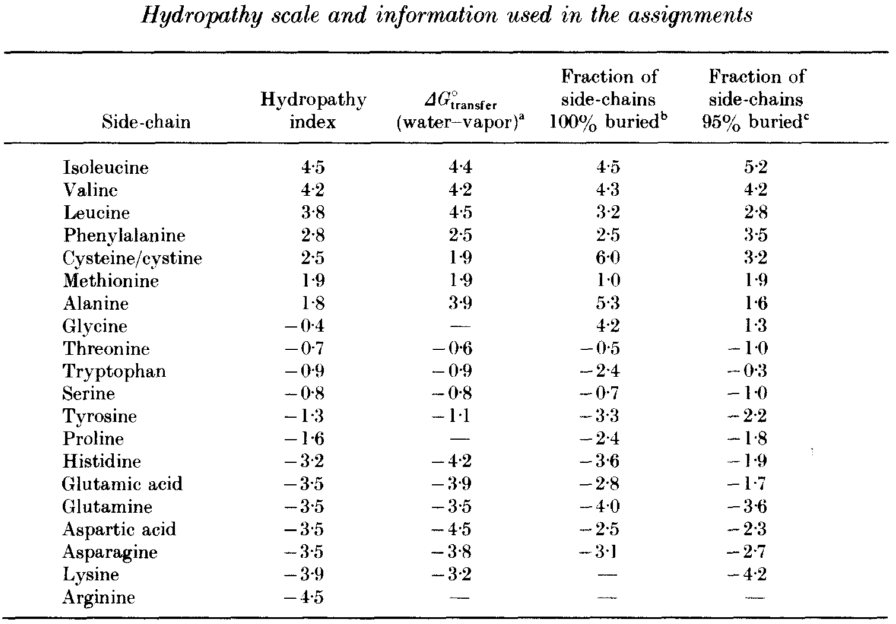
Proteins, though built from amino acids, have both basic and acidic properties (also called Zwitterionen). This comes from the various side chains of amino acids having various \(pK_a\) values. The acid dissociation constant \(K_a\) is commonly given in a negative logarithmic scale \(pK_a\) due to the many orders of magnitude spanned. The larger \(K_a\) is, the more dissociation is taking place, thus the stronger the acid in solution (more protons donated following the Brønsted-Lowry scheme).
Like this, proteins can be separated according to their isoelectric point using a technique called isoelectric focusing. It uses a \(pH\) gradient to separate proteins on a gel, and also the first step in 2-D gel separation.
The \(pI\) of a protein can be calculated or experimentally determined with isoelectric focusing.
The theoretical calculation can be done by considering all free ionizable groups. These are the N-terminal amino group, the C-terminal carboxyl group and the ionizable sidechains:
- Asp (D)
- Glu (E)
- Arg (R)
- Lys (K)
- His (H)
- Cys (C) (-SH)
- Tyr (Y)
Histidine (H), lysine (K), and arginine (R) carry a proton acceptor group in their side chains.
Cysteine (C), aspartate (D), glutamate (E), and tyrosine (Y) have proton donor groups in their side chains.
Note that the \(pK\) values for the termini also vary with the actual amino acids that form the corresponding ends.
Using the dissociation constants for these ionizable groups we can calculate a estimation of the pI of a protein. The partial charges are calculated for each amino acid x at any given \(pH\) is:
\(C_x = 1/(1 + 10^{pH-pK_x})\)
if the amino acid is positively charged, and
\(C_x = -1/(1 + 10^{pK_x-pH})\)
if the amino acid is negatively charged.
A function \(C_P(pH)\) can now estimates the net charge of a protein \(P\) for a given \(pH\) with:
\(C_P(pH) = \sum_{x in P}{C_x}\)
The protein's \(pI\) is \(C_P(pH) = 0\).
Here is an algorithm that calculates the pI of a given sequence of amino acids:
const: \(\epsilon\) limit for accuracy
var: \(x,x_0,x_1\)
begin- find values \(x_0, x_1\) that \(sign(C(x_0)) = sign(C(x_1))\)
- while [\(x_1 - x_0\)] \(\geq \epsilon\) and max number of iterations not reached do
- \(x := (x_0 + x_1)/2\)
- if \(sign(C(x_0)) \neq sign(C(x))\) then \(x_1 := x\) else \(x_0 := x\) end
- end
end\(pI\)
is in [\(min(x_0, x_1), max(x_0, x_1)\)]
EXERCISE: calculate the isolelectric point of the peptide SYFPEITHI.